Integrated Genomics Analysis
Comprehensive Integrated Analysis
While accurate sequencing data is important, advanced tools for sequence alignment, variant calling and database integration are crucial to translate genomics information into actionable interpretation. ACT Genomics integrates advanced annotation and visualization tools, coupling with public and proprietary databases to deliver comprehensive and comprehensible reports.
In addition to collecting data from multiple public genomic databases, ACT Genomics also conducted an in-house sequencing project to establish baseline data for technical noise filtering and polymorphism annotation. Through these efforts, ACT Genomics has successfully improved the accuracy of variant calling and annotation.
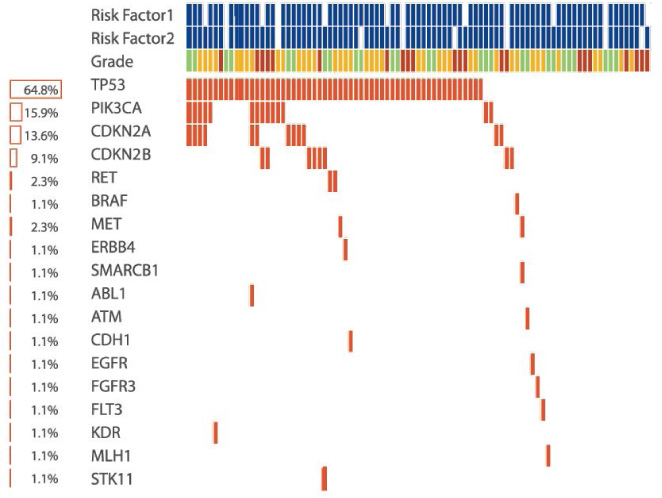
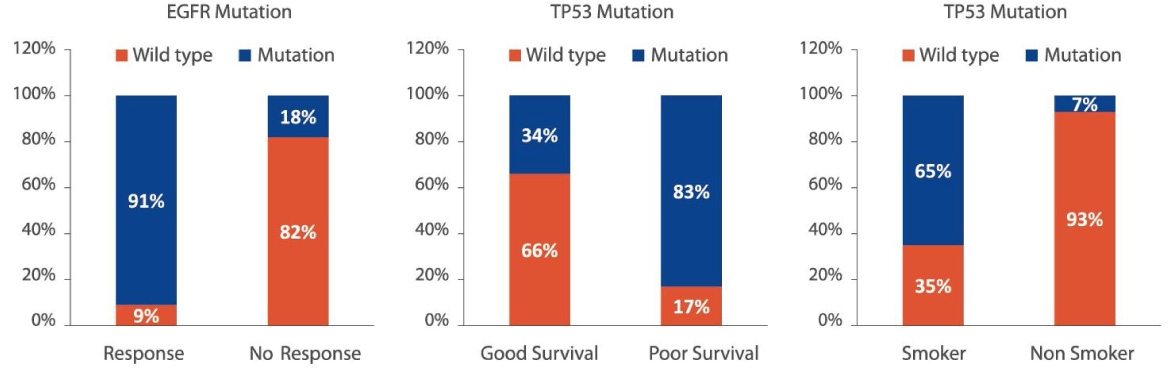
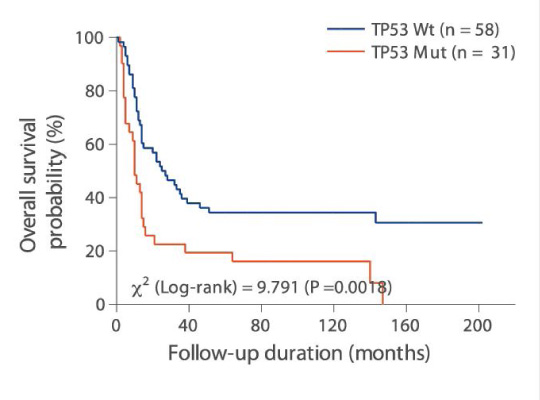
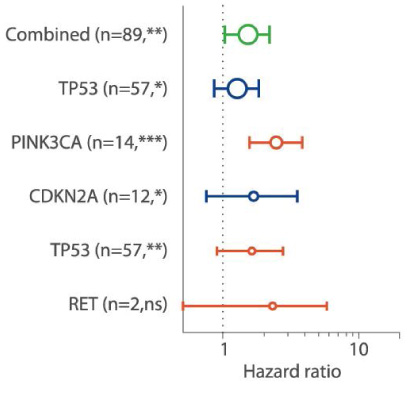
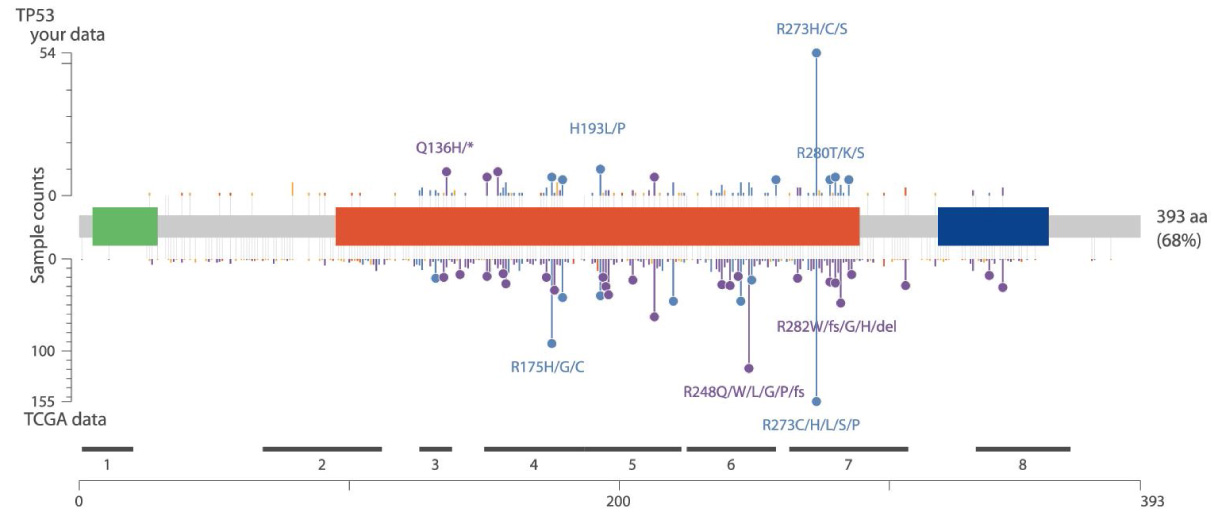
For clinical research projects, ACT Genomics provides integrated sequencing QC results, project-level variant summary, distribution pattern of non-synonymous mutations, as well as comparison of mutation spectrum of 5 selected genes with TCGA data as standard project report. To facilitate the research and publication process, additional statistical analysis such as group-wise variant comparison and survival analysis, including Kaplan-Meier analysis and Cox regression analysis, will also be performed if clinical data is provided.